Bioinformatics is (broadly speaking) the application of information technology to biological problems. This term appeared for the first time in 1970 in an article written in Dutch, where it was proposed as “the study of computer processes in biotic systems”, a meaning different from that of today although, in some fields more theorists of modern bioinformatics, this definition remains valid.
In 1955 Frederick Sanger published the amino acid sequence of insulin, the first sequence of a protein to be discovered. This fundamental work (for which Sanger received the Nobel Prize for chemistry in 1958), paved the way for protein sequencing. The sequencing technology, initially manual, was improved until it was fully automated by Pehr Edman in 1967. The fact that the primary structure of proteins consisted of unique sequences of amino acids was in itself an IT concept. The technology of protein sequencing and the consequent growth of the number of available sequences created computational needs:
High molecular weight protein sequencing involved the partial enzymatic digestion of proteins into peptides that were sequenced. This strategy consequently required the correct assembly of the partial sequences in a single final sequence.
The comparison of sequences of homologous proteins, that is belonging to different species descending from a common ancestor for the creation of phylogenetic trees.
At the same time computers were beginning to be available in the most advanced research centers in the USA, and their programming had been simplified thanks to the FORTRAN language (introduced by IBM in 1957). Already in the mid-1960s, Cyrus Levinthal and his group first used a computer at MIT to build a 3 D model of cytochrome C.
Some pioneers of bioinformatics, including Margaret Dayhoff and Walter Fitch, compiled the first programs for the computerized execution of the assembly of protein sequences and the comparison between sequences and the creation of phylogenetic trees.
In 1970 Saul Needleman and Christian Wunsch perfected the comparison between two sequences with the publication of an innovative algorithm for the analysis of similarities.
DNA sequencing, invented in 1977 by Allan Maxam and Walter Gilbert and perfected by Frederick Sanger, gave rise to an exponential production of gene sequences, giving further impetus to bioinformatics. The representation of DNA and protein sequences as character strings was ideal for their computerized manipulation.
Programs were created for storing sequences as digital files, for printing them, for identifying sites of restriction enzymes or sequences coding within DNA sequences, or for translating DNA sequences into sequences of amino acids. The exponential growth of DNA and protein sequences led to the creation of programs, such as BLAST, capable of rapidly comparing an unknown sequence with a bank of known sequences.
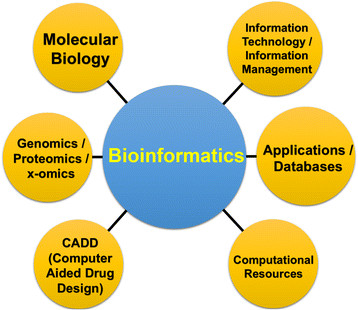
It is not possible to summarize here the enormous development of bioinformatics in the last 35 years, but it is enough to say that it has grown in parallel with the immense advances in molecular biology, genetics and protein biochemistry, as well as, of course, the progress of computer science and computers. Modern bioinformatics is divided into three main fields: *